Implen Journal Club | February Issue Method of the Year 2022 issue |
|
This Month’s Implen NanoPhotometer® Journal Club: Method of the Year 2022 Edition is highlighting long-read sequencing, which was named Nature’s Method of the Year 2022 given its instrumental role in the complete sequencing of a human genome being published in Science. The first week's issue is discussing the use of long-read sequencing in the study of one of the best potential sources of spices from Indonesia, the tropical fruit tree species called Myristica fatua having very few genomic resources available for the species. Matra et al. submitted, in the journal of Data in Brief, the Myristica fatua coding sequence (CDS) as the first transcriptome reference with a full-length transcriptome assembly obtained utilizing long-read sequencing from Oxford Nanopore Technologies. Full-length transcripts can be obtained from this data, which is helpful for researching gene expression analyses. This information offers datasets of EST-microsatellite molecular markers for crop breeding programs in genera related to Myristica fatua, as well as the potential to identify full-length transcripts involved in flavonoid biosynthesis for use in downstream analysis in Myristica fatua and related genera by molecular biologists. The NanoPhotometer® NP80 was used in this study to assess the quality and quantity of RNA extracted from the Myristica fatua leaves. |
|
Next issue is highlighting an NAR breakthrough article published by Christopher Faulk illustrating the time and cost saving potential of long-read sequencing- Nature’s Method of the Year 2022. Typically, reference animal genome generation is a time-consuming and costly operation. DNA sequencing using nanopore technology provides direct, real-time, long reads, scalable, portable, automated, rapid and comprehensive genomic analysis. Faulk sequenced the genome of the black carpenter ant for only $1000 as a sole researcher in just one week. Along with the nuclear genome, the mitochondrial genome was assembled along with the mitochondrial genome and two commensal bacteria species living within the ant. Nanopore technology also enabled epigenetic measurements from the same ant and replicated other studies showing very low DNA methylation. The reference genome compared favorably to other ant species in continuity and protein prediction accuracy. This method will allow other low-resource labs to create high quality genome assemblies with a low cost. The NanoPhotometer® N60 was used in this study to assess the quality of the purified DNA. |
|
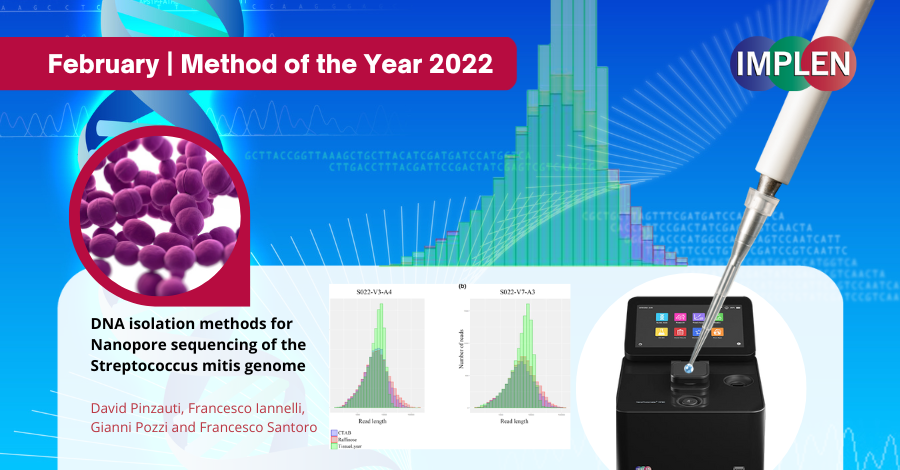
Next issue is exploring the possibility of achieving long-sequencing reads that makes Oxford Nanopore technology an essential tool for bacterial genomics. Streptococcus mitis is a Gram-positive bacterium, member of the oral commensal microbiota. It can occasionally be the etiologic agent of diseases such as infective endocarditis, bacteraemia and septicaemia. The repetitive nature of the S. mitis genome impairs assembly of a complete genome relying only on short sequencing reads. Oxford Nanopore long-read sequencing was shown to overcome this limitation by generating long reads, enabling the resolution of genomic repeated regions and the assembly of a complete genome sequence. Since the output of a Nanopore sequencing run is strongly influenced by genomic DNA quality and molecular weight, the DNA isolation is the crucial step for an optimal sequencing run. Pinzauti et al. recently published the results of the comparison of three DNA isolation methods, which were validated for the sequencing of the highly recombinogenic S. mitis genome. The methods aimed to isolate a pure high molecular weight DNA from S. mitis to be used as a template in Oxford Nanopore sequencing experiments. The findings demonstrated that sequencing of DNA isolated with a mechanical lysis-based method, despite being cheaper and quicker, did not generate ultra-long reads (maximum read length of 59516 bases) and did not allow the assembly of a circular complete genome. The DNAs isolated using the two enzymatic lysis-based methods generated ultra-long reads, up to 181199 bases, achieving circular complete genome assembly. These methods can be readily applied for isolation of high molecular weight genomic DNA from difficult to lyse Gram-positive bacteria. The NanoPhotometer® was used in this study to determine the genomic DNA purity grade evaluating the absorbance ratios 260 nm/280 nm and 260 nm/230 nm. |
|
The final issue is highlighting the work involving Amazon parrots in the Greater Antilles representing a fascinating model of speciation on islands, similar to Darwin's finches in the Galapagos. Amazon parrots colonized the islands of the Greater Antilles from the Central American mainland, but there was no consensus as to how and when this happened. Kolchanova et. al. recently published in the journal of genes, a study that sequenced with long-read sequencing and annotated full mitochondrial genomes of all the extant Amazon parrot species from the Greater Antillean (A. leucocephala, A. agilis, A. collaria, A. ventralis, and A. vittata) including annotated mitogenome maps to provide information on sequence organization, variation, population diversity, and evolutionary history. The data from this study support the stepping-stone dispersal and speciation hypothesis that has started approximately 3.47 MYA when the ancestral population arrived from mainland Central America and led to diversification across the greater Antilles, ultimately reaching the island of Puerto Rico 0.67 MYA. This analysis contributes to understanding evolutionary history and empowers subsequent assessments of sequence variation and helps design future conservation efforts. The NanoPhotometer® C40 was used in this study to measure the extracted genomic DNA. |
©2023 Implen. All rights reserved.