Implen Journal Club | January Issue |
|
Innovations in Nanopore Sequencing: A Novel Label-Free Approach for Small Molecule QuantificationThis New Year’s Implen NanoPhotometer Journal Club is highlighting an article published in 2024 by Quint et. al. that introduced a novel label-free method for quantifying small molecules using the nanopore sequencing platform, focusing on ethanolamine as a model analyte. The novel method involved a strand displacement assay, where the target-binding aptamer is displaced by ethanolamine, and the non-displaced aptamer is detected by a nanopore system, allowing quantification in the micromolar range. Ethanolamine is important for healthy cell membranes and metabolism. Decreased levels are linked to neurological diseases and cancer. Ethanolamine also regulates virulence in pathogenic gut bacteria. Therefore a simple detection method could help study these diseases and processes. This label-free approach utilizes protein nanopores for detecting small molecules and is significant for its potential applications in clinical diagnostics and drug development. This study also introduced Nanotrace, a new visualization and analysis software. This approach demonstrates protein nanopores, like those in a low-cost sequencer from Oxford Nanopore, can quantitatively detect small molecules using target-specific aptamers without labels. The novel approach presented in this research provides a promising alternative for small molecule detection in various fields, including environmental chemistry, life sciences, and disease biomarker identification. The NanoPhotometer® was used in this work to determine the amount of immobilized aptamer. The same measurements were done with non-reacted oligonucleotides as a control to the total absorption. This gave insights on the ability of the aptamer to hybridize to its complementary strand and the effect of steric hindrance. |
|
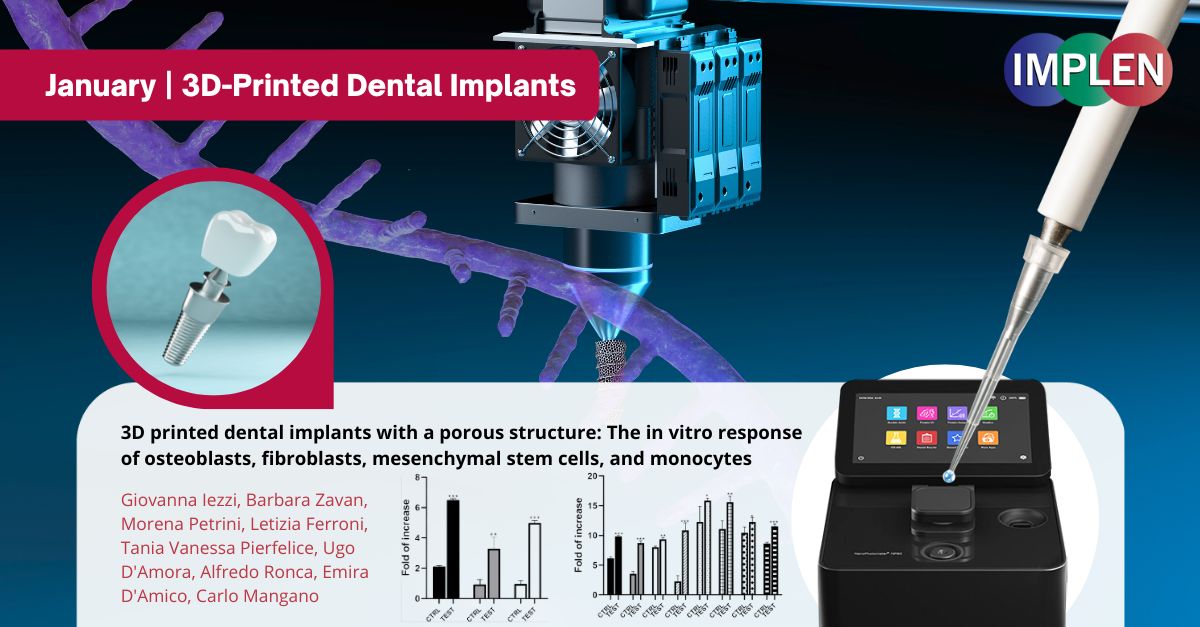
Revolutionizing Dental Care: The Promise of 3D-Printed Porous Dental ImplantsNext, Implen NanoPhotometer Journal Club is sharing exciting news in dental technology with a groundbreaking study by Iezzi et al. published in 2024 in the Journal of Dentistry that has been conducted on 3D-printed dental implants, revealing some impressive findings. These implants, created through selective laser melting and organic acid etching, have a unique porous structure, resulting in a design that significantly enhances cell adhesion and growth, and expression of osseointegration biomarkers. This study "3D printed dental implants with a porous structure: The in vitro response of osteoblasts, fibroblasts, mesenchymal stem cells, and monocytes" characterized the implant’s surface topography and investigated the biological response with how these implants interact with important cell types. This study employed various methodologies, including scanning electron microscopy (SEM) analysis, cell culture, and multiple assays to assess cell proliferation, attachment, gene expression, mineralization, and collagen production. The promising results demonstrated better cell attachment and growth, which is crucial for successful dental procedures. This could be a major advancement in dental implantology, potentially improving patient care and success rates in dental treatments. The Implen NanoPhotometer NP80 spectrophotometer was used in this study to quantify total RNA. This step was important for evaluating gene expression in human oral osteoblasts (hOBs) and gingival fibroblasts (hGFs) using RT-qPCR. Specifically, RNA was isolated and its concentration measured using the NP80, which provided the necessary data for subsequent cDNA synthesis and gene expression analysis. #NanoPhotometer #NP80 #Implen #DentalInnovation #3DPrinting #HealthTech |
|
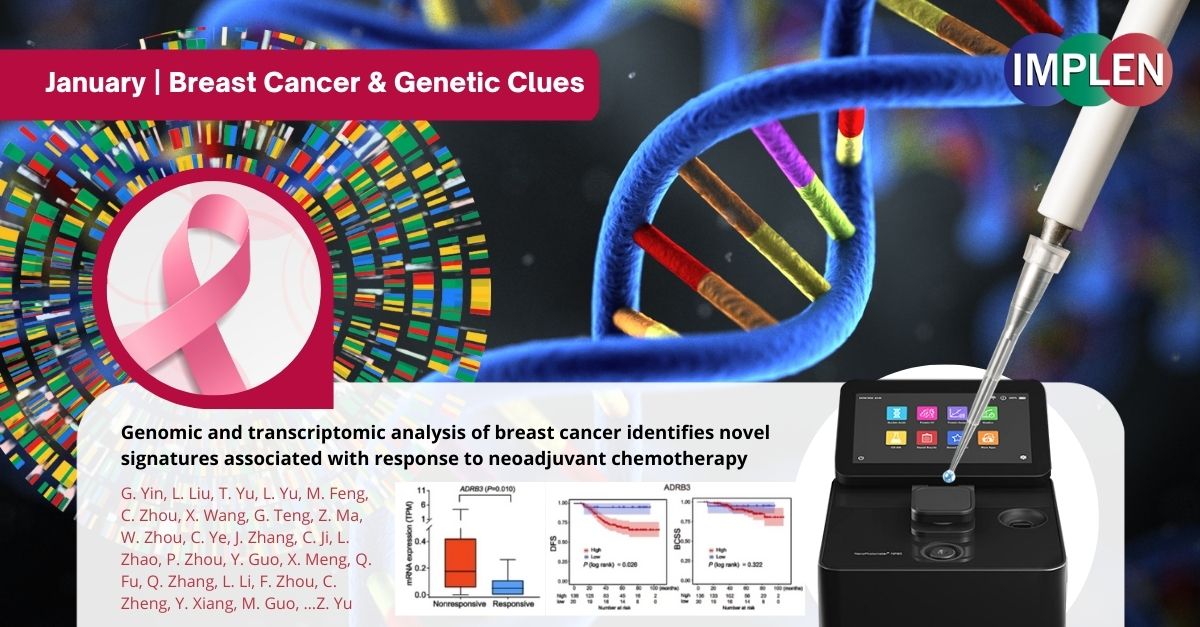
Uncovering Genetic Clues to Breast Cancer Chemo Response: Study Finds Mutations and Gene Expression Changes Linked to Neoadjuvant Treatment EfficacyThis issue is highlighting the work published in 2024 by Yin et. al. using Illumina’s NovaSeq technology to uncover molecular signatures of breast cancer tumors associated with chemotherapy response. Key findings of this study indicate neoadjuvant chemotherapy significantly alters mutation rates, DNA repair pathways, and immune microenvironment. Specifically, the CDKAL1P409L mutation was identified as reducing chemotherapy sensitivity, while amplifications in ADGRA2 or ADRB3 were linked with worse prognosis and treatment outcomes. This comprehensive analysis of genomic and transcriptomic changes in breast cancer tumors following neoadjuvant chemotherapy treatment reveals insights into the impact of chemotherapy on the molecular landscape of breast cancer. The findings highlight potential genetic biomarkers that provide understanding into chemotherapy resistance mechanisms and hold promise for guiding more personalized and effective treatments based on a patient's genetic profile. This study provided insights into breast cancer tumor adaptation and treatment response, paving way for genetics-guided improvements in prognostic predictions and treatment strategies. The NanoPhotometer® was used in this study for the quantification of DNA and RNA. Specifically, after isolating total DNA from fresh frozen tissue samples and blood samples, and extracting RNA from fresh frozen tumor tissue, the purity of the total DNA and RNA was estimated using the NanoPhotometer. |
|
Revolutionizing Soil Remediation: Laccases in the Degradation of Petroleum HydrocarbonsThe last issue is showcasing the newly published study in 2024 by Diefenback et. al. titled "Laccase-mediated degradation of petroleum hydrocarbons in historically contaminated soil", which investigated the use of laccases, specific enzymes, for the bioremediation of petroleum hydrocarbons in polluted soils. Highlighting the limitations of traditional remediation methods, this research turned to bioremediation as a cost-effective, environmentally friendly alternative. This study successfully isolated novel laccases from local microorganisms at a polluted site in Droesing Austria. Among these, the laccase PlL, derived from Parvibaculum lavamentivorans, stood out for its ability to significantly reduce petroleum hydrocarbons, especially larger molecules. This enzyme not only outperforms conventional microbial treatments but also demonstrates superior efficiency compared to other laccases like MtL from Myceliophthora thermophila. This study has shown laccase-mediated bioremediation, especially using PlL, as a promising approach for managing petroleum hydrocarbon pollution, especially in areas with long-standing contamination. This method offers a more efficient, environmentally friendly remediation, potentially reducing the time and complexity associated with traditional methods. The NanoPhotometer® was used in this study to assess the quality and quantity of extracted gDNA. Additionally, the concentration of the purified proteins was determined using a NanoPhotometer. |
©2024 Implen. All rights reserved.